MWD: General description¶
Purpose¶
Application to analyze Molecular Weight Distributions
Data Files¶
The first line of the file should contain the sample parameters separated by semi-colons (
;). It may contain any number of parameters which will be read and saved as file-parameter in RepTate.Then the data columns should appear, separated by spaces or tabs.
.gpc extension¶
Text files with .gpc extension should be organised as follows:
.gpcfiles should provide parameter values fornumber-average molar mass, \(M_n\)
weight-average molar mass, \(M_w\)
polydispersity index, \(\text{PDI}\)
2 columns separated by spaces or tabs containing respectively:
molar mass, \(M\),
weight associated, \(\dfrac{\text d w(\log M)}{\text d \log M}\).
A correct .gpc file looks like:
Mn=100;Mw=250;PDI=2.5
0.1148E+03 0.5000E-03
0.1514E+03 0.1500E-02
0.1995E+03 0.1667E-02
0.2630E+03 0.1750E-02
0.3467E+03 0.5250E-02
0.4571E+03 0.6750E-02
... ...
.reac extension¶
Text files with .reac extension should be organised as follows:
.reacfiles are not expected to provide any specific parameter values.4 columns separated by spaces or tabs containing respectively:
molar mass, \(M\),
weight associated, \(\dfrac{\text d w(\log M)}{\text d \log M}\),
the g-factor, \(g(M)\),
number of branch per 1000 carbon, \(\log_{10}(g(M))\).
A correct .reac file looks like:
0.1148E+03 0.5000E-03 0.1000E+01 0.0000E+00
0.1514E+03 0.1500E-02 0.1000E+01 0.0000E+00
0.1995E+03 0.1667E-02 0.1000E+01 0.0000E+00
0.2630E+03 0.1750E-02 0.1000E+01 0.0000E+00
0.3467E+03 0.5250E-02 0.1000E+01 0.0000E+00
0.4571E+03 0.6750E-02 0.9977E+00 0.3765E+00
... ... ... ...
Views¶
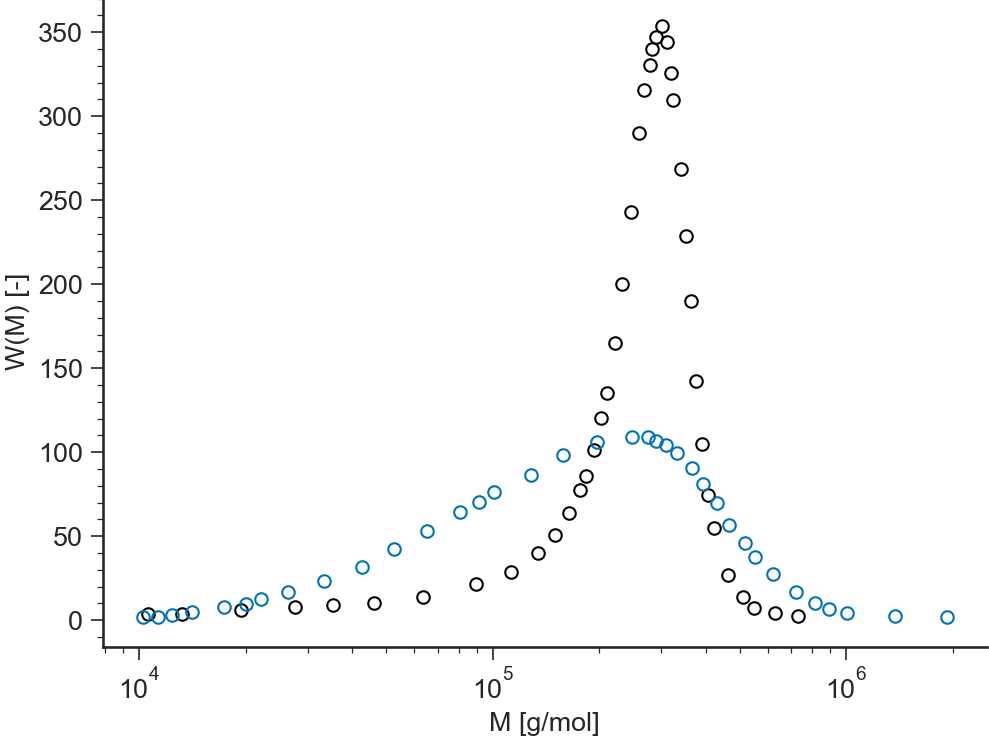
log-lin¶
(Molecular weight in logarithmic scale)
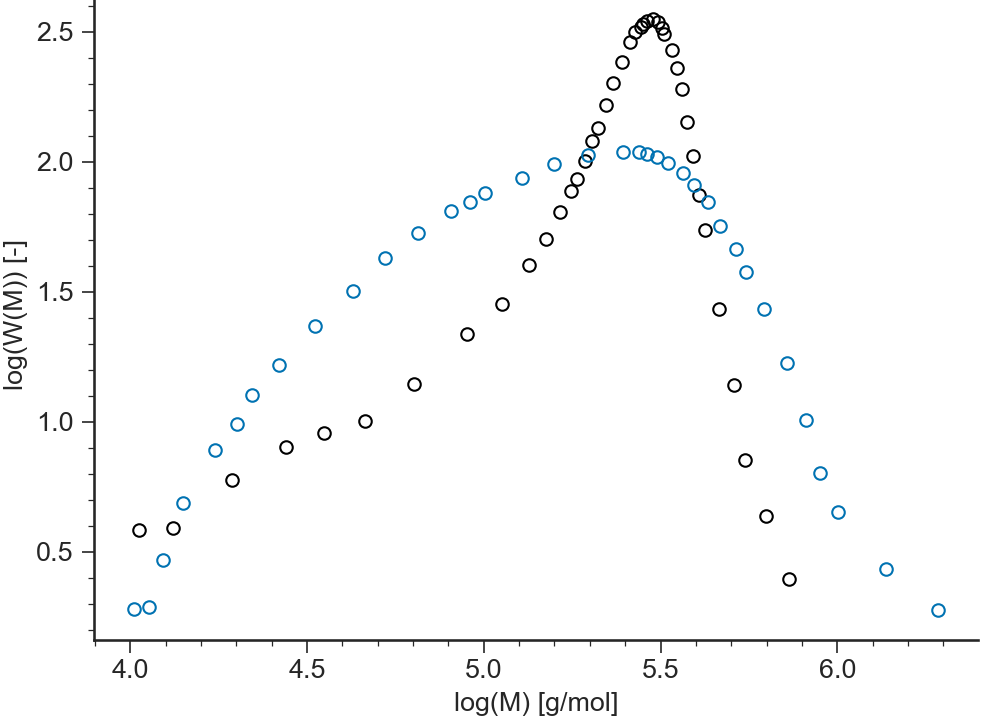
log-log¶
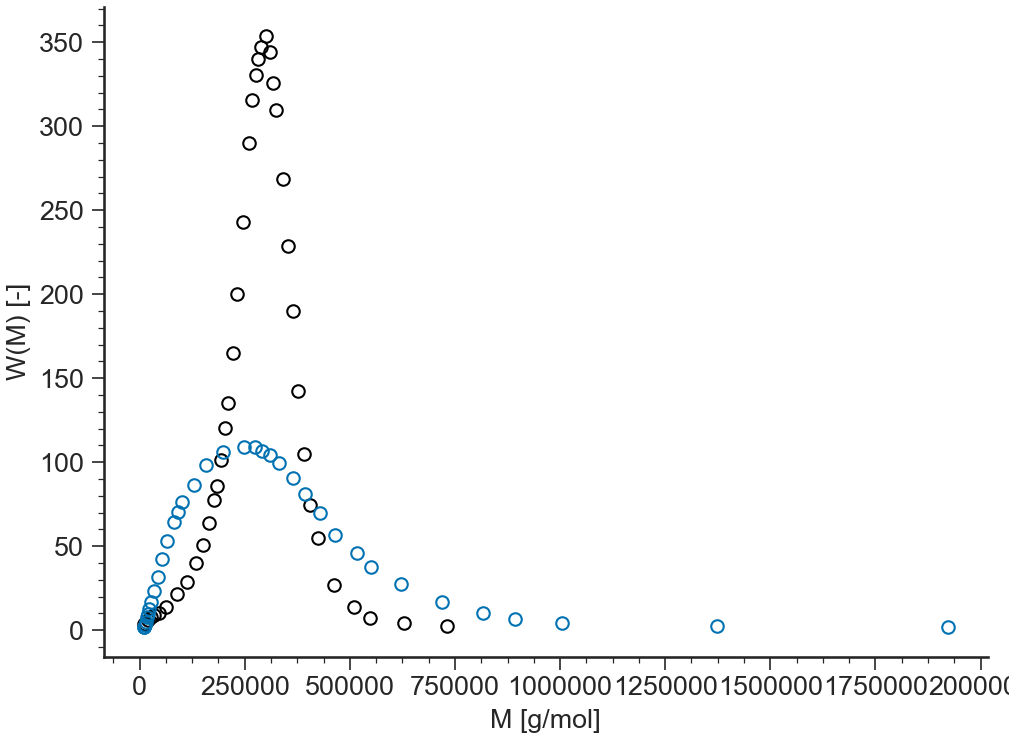
lin-lin¶