Creep: General description¶
Purpose¶
Application to Analyze Data from Creep experiments
Data Files¶
The first line of the file should contain the sample parameters separated by semi-colons (
;). It may contain any number of parameters which will be read and saved as file-parameter in RepTate.Then the data columns should appear, separated by spaces or tabs.
.creep extension¶
Text files with .creep extension should be organised as follows:
.creepfiles should contain at least the parameter value for the:Applied stress
stress
2 columns separated by spaces or tabs containing respectively:
time, \(t\),
strain, \(\gamma\),
A correct .creep file looks like:
stress=10;chem=PE;label=CM3;T=150;
t strain stress T
s - Pa C
1E-3 1.413E-5 10 149.9
1E-3 -9.419E-6 10 149.9
1E-3 -2.826E-5 10 149.9
... ... ... ...
Views¶
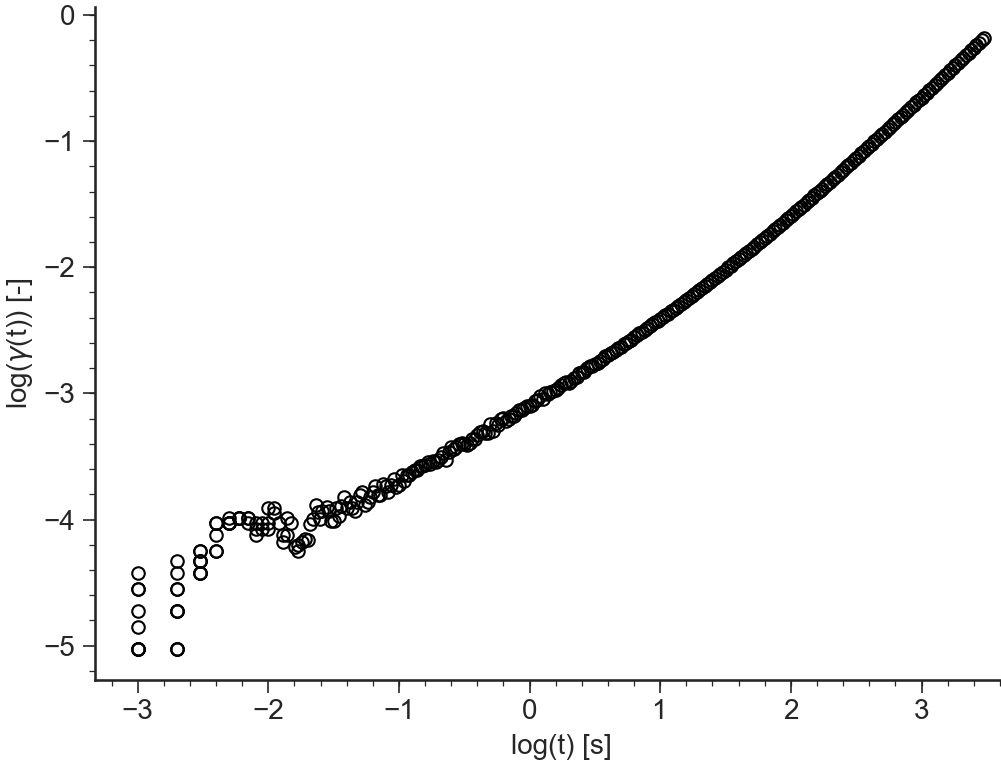
log(gamma(t))¶
- ApplicationCreep.viewLogStraint()[source]¶
Logarithm of the applied strain \(\gamma(t)\) vs logarithm of time \(t\)
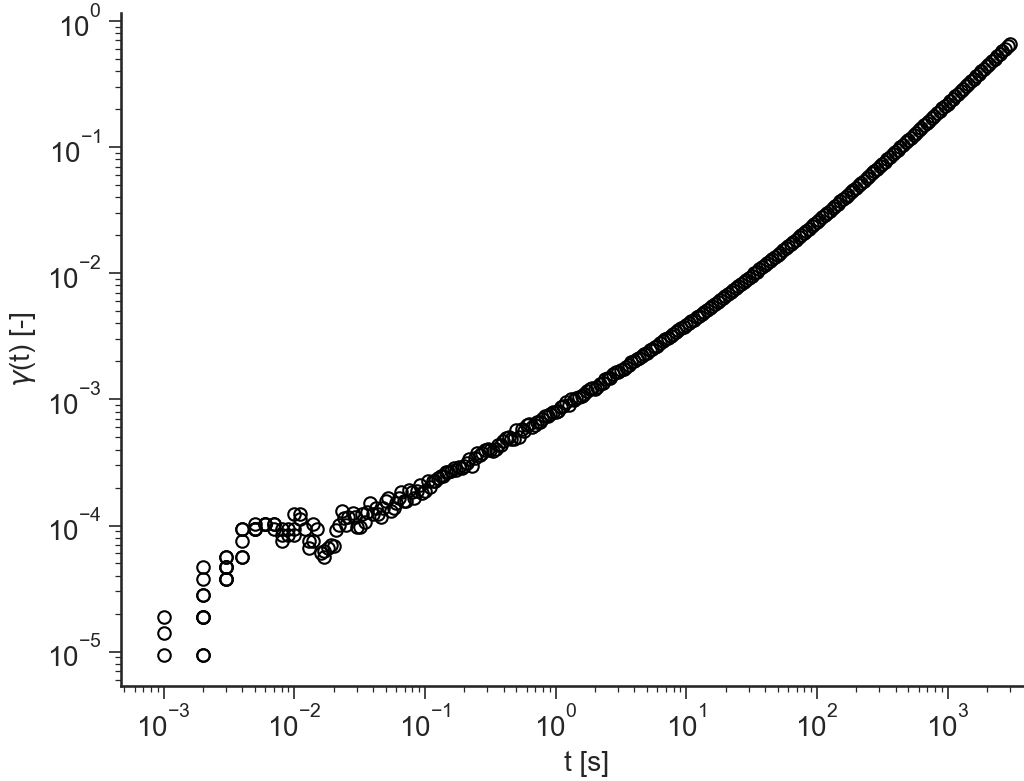
gamma(t)¶
- ApplicationCreep.viewStraint()[source]¶
Applied strain \(\gamma(t)\) vs time \(t\) (both axes in logarithmic scale)
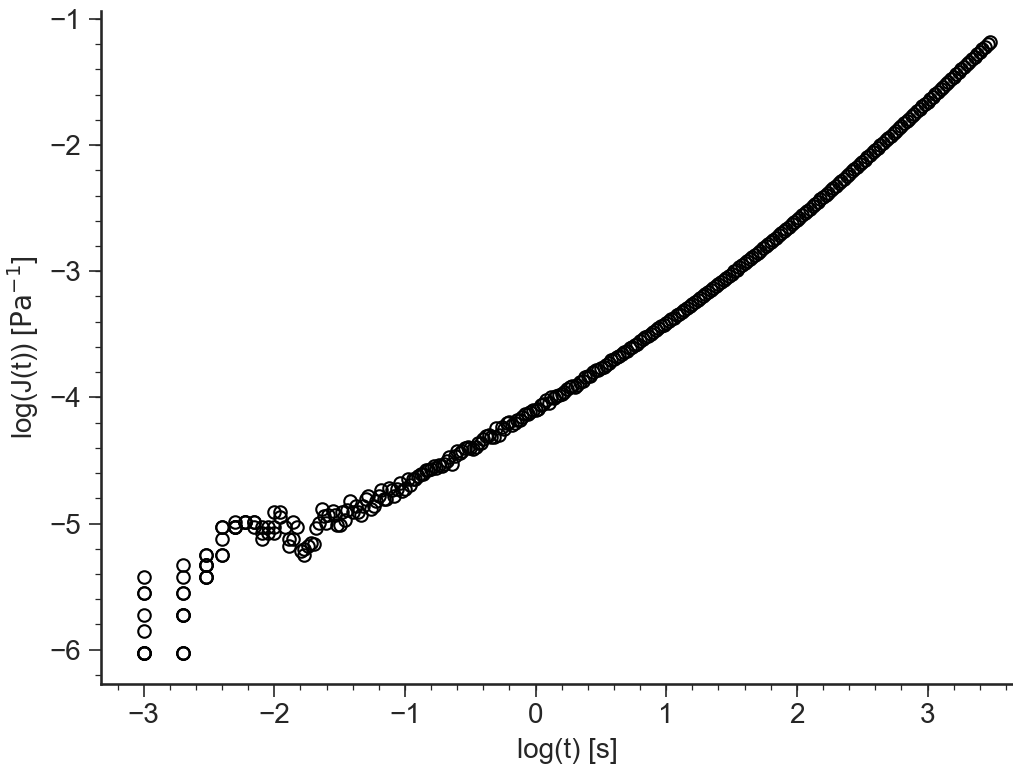
log(J(t))¶
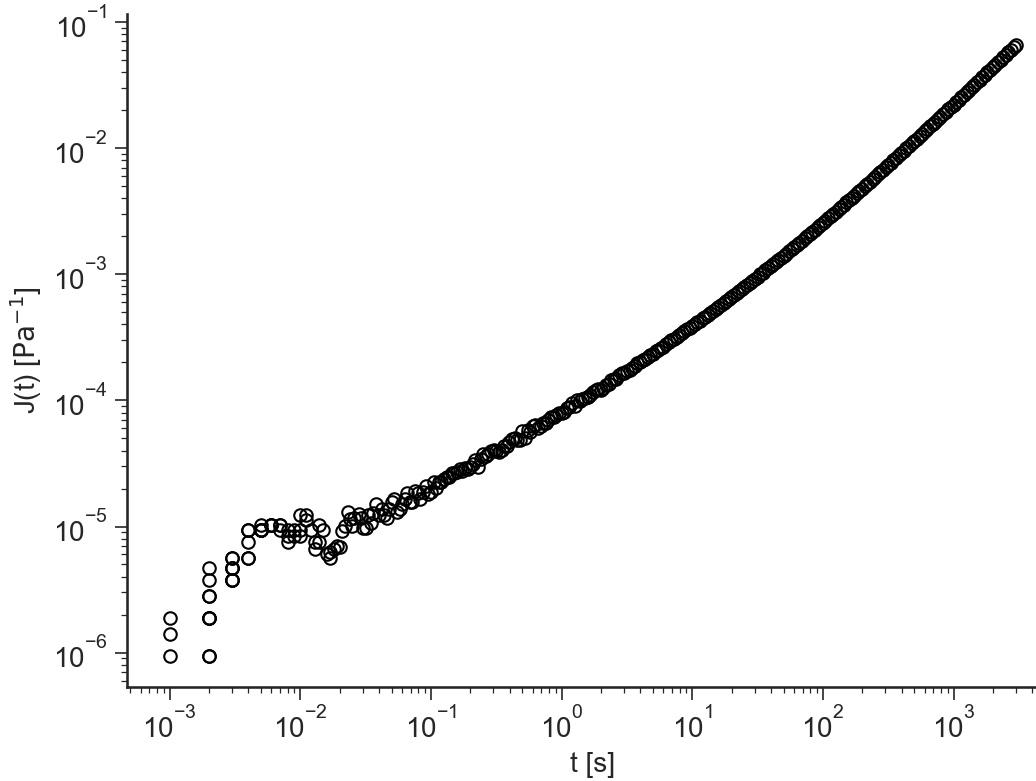
- ApplicationCreep.viewLogJt()[source]¶
Logarithm of the compliance \(J(t)=\gamma(t)/\sigma_0\) (where \(\sigma_0\) is the applied stress in the creep experiment) vs logarithm of time \(t\)
J(t)¶
- ApplicationCreep.viewJt()[source]¶
Compliance \(J(t)=\gamma(t)/\sigma_0\) (where \(\sigma_0\) is the applied stress in the creep experiment) vs time \(t\) (both axes in logarithmic scale)
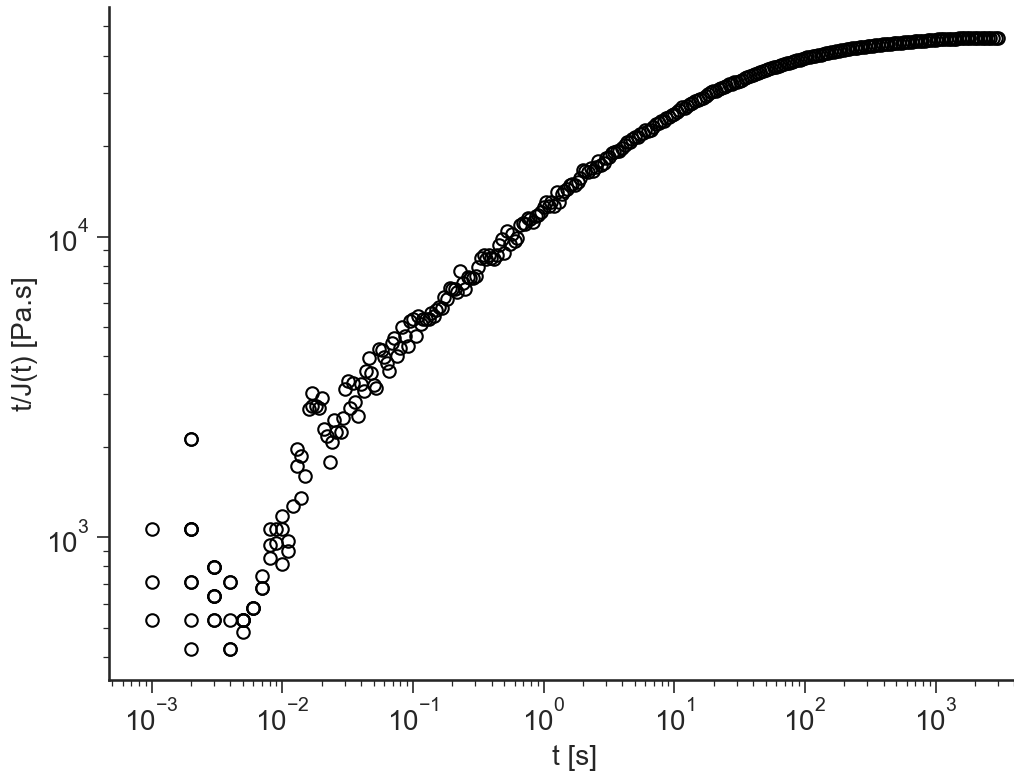
t/J(t)¶
- ApplicationCreep.viewt_Jt()[source]¶
Time divided by compliance \(t/J(t)\) vs time \(t\) (both axes in logarithmic scale)